GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
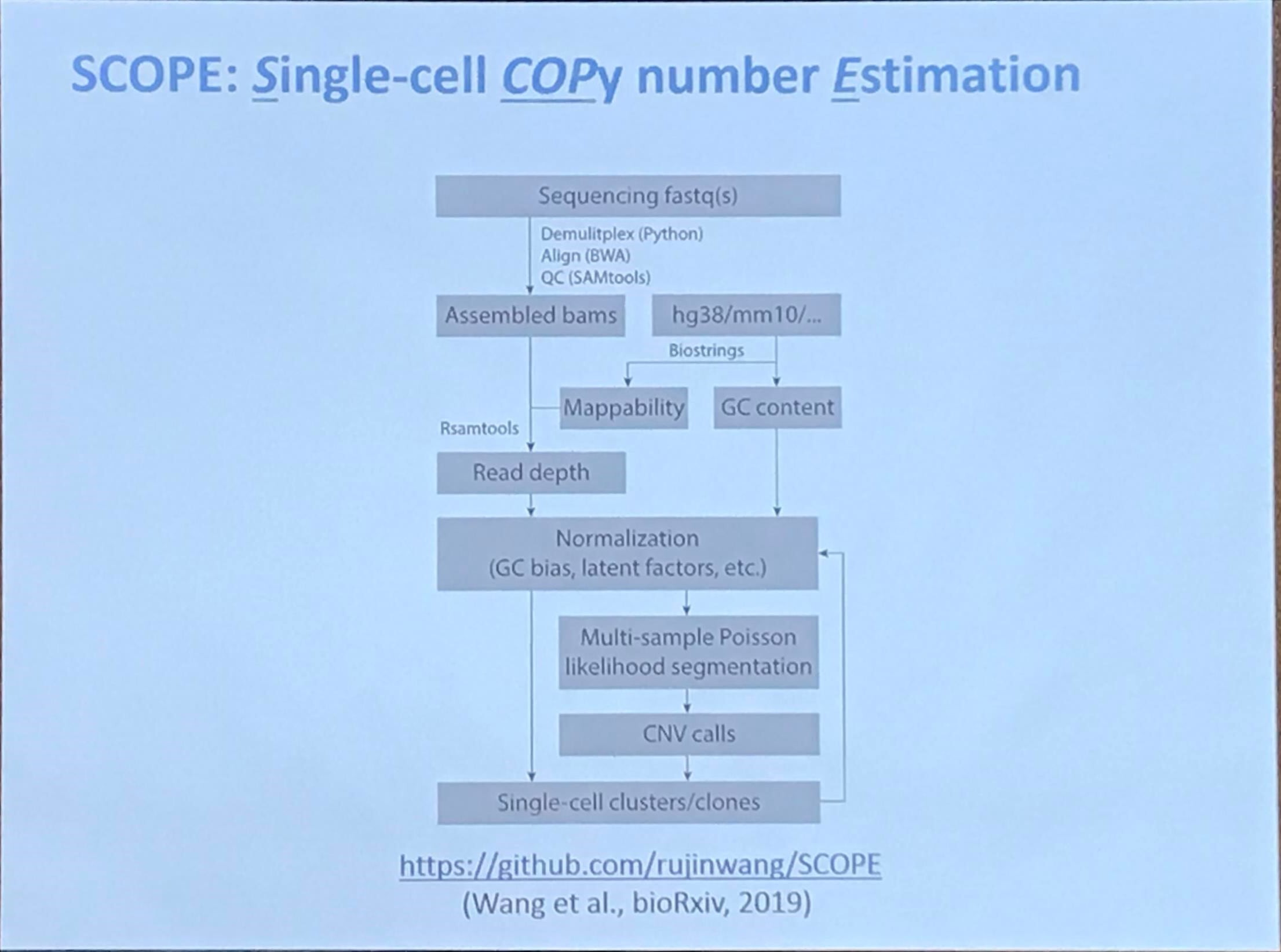
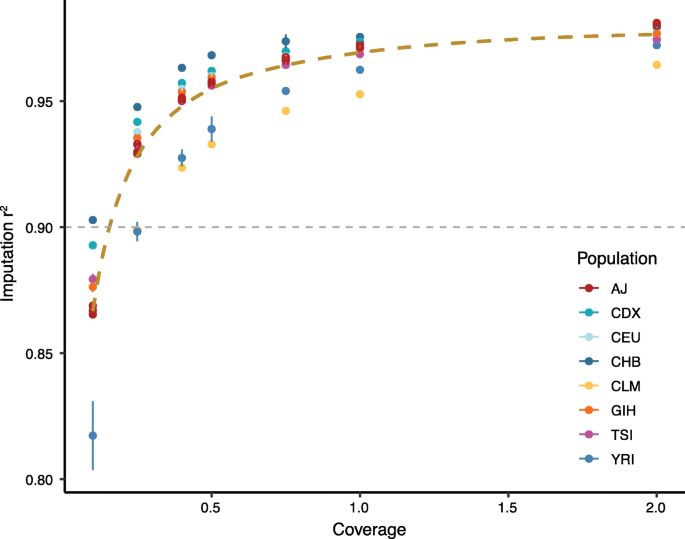
DNA copy number profiling: from bulk tissue to single cells

PDF) Low-Coverage Whole Genome Sequencing Using Laser Capture Microscopy with Combined Digital Droplet PCR: An Effective Tool to Study Copy Number and Kras Mutations in Early Lung Adenocarcinoma Development

GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data
Bamgineer: Introduction of simulated allele-specific copy number variants into exome and targeted sequence data sets
GitHub - AdelmanLab/GetGeneAnnotation_GGA
awesome-omics/GENOMICS.md at main · flowhub-team/awesome-omics · GitHub
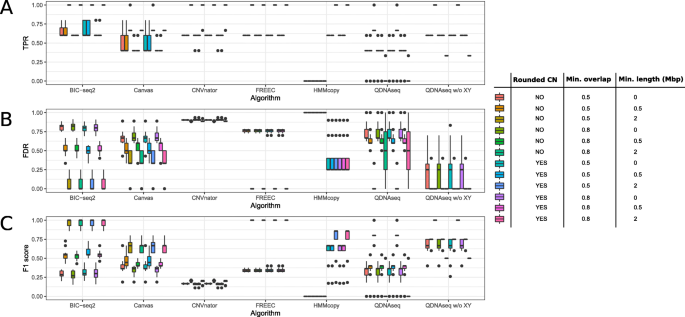
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics

CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing, Genome Biology
jpoell · GitHub

Detection of Copy Number Variation using Shallow Whole Genome Sequencing Data to replace Array-Comparative Genomic Hybridization Analysis